Hierarchical Active Transfer Learning
In my last post, we explored a machine learning method known as transfer learning used to counter problems in which two models have similar purposes but data is more readily available for one than the other. By exploiting similarities in data, transfer learning allows us to leverage one model to increase accuracy in the other, and vice versa. Most commonly this is done by substituting a series of layers in one model with the others, but in this post we will discuss a model-independent framework known as Hierarchical Active Transfer Learning (HATL) [1].
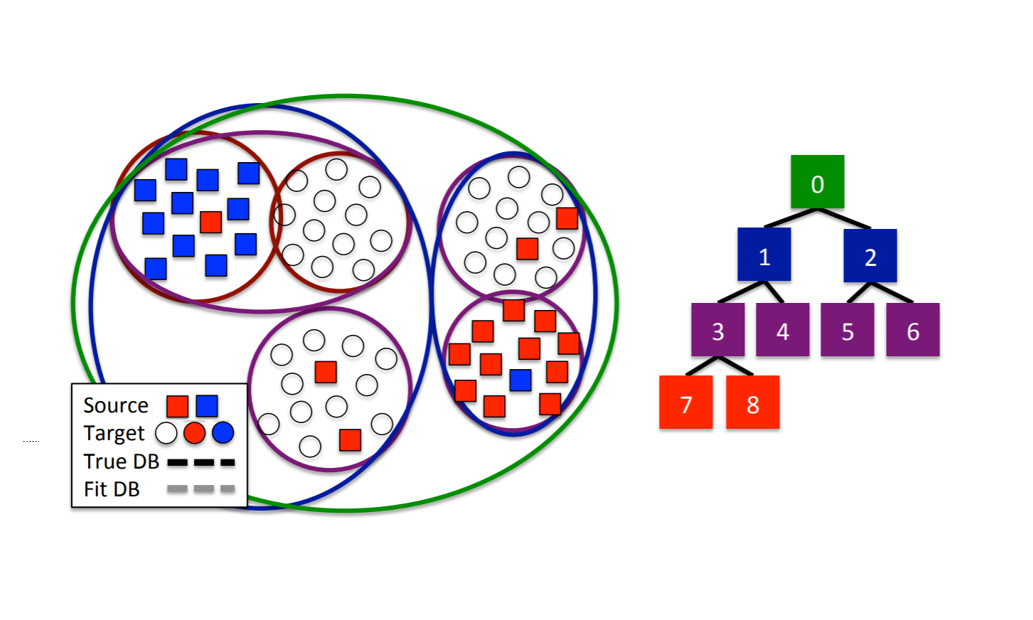
Notation: Here we will establish the task for which we want to leverage labeled data from as the source domain, while the task for which we want to use said leveraged labeled data is referred to as the target domain.
HATL utilizes both transfer learning and active learning, and before we explore deeper we should establish the pitfalls and advantages of both. We first note that labeled data is scarce and unlabeled data is plentiful. As a brief summary to any unfamiliar, active learning is a machine learning method that allows one to “smartly query” unlabeled data points in such a way that the unlabeled data that is queried is as helpful as possible for our model to learn from. In many applications, it is often costly to label data, whether that cost is time or money. e.g. using RNA-seq for transcriptomics analysis – active learning reduces the amount of labeled data needed. It however also suffers from a circular dependency on data – in order to pose good queries, we need to have a good model, and in order to have a good model we need to have labeled data. Transfer learning, on the other hand allows for source domain knowledge to provide an initial bias for the target domain, which can later be adapted. e.g. gene expression prediction in one omic using weights of a model that does gene expression in another, similar omic. Transfer learning suffers from negative transfer – due to the source and target domains being too different, knowledge transferred from the source actually detriments the target. It is apparent that active learning and transfer learning have complementary strengths – active learning can be applied to tell us which data to label such that when transfer learning is applied prediction results are optimal. Pitfalls are also somewhat addressed by integrating the two, but rely heavily on how similar data from the source and target domain are. With this in mind, we are now ready to explore the HATL algorithm.
Introduced by Kale et. al. in 2015, HATL ties active learning and transfer learning by clustering. Using hierarchical clustering, the source and target data are initially clustered into a tree and fed as input to the algorithm. A list of pruned nodes of the tree is initialized to the root and will be used to keep track of the optimal clustering of the data (each node in the tree refers to a cluster via our hierarchical clustering). The algorithm first labels only the source data as a starting point for active learning to make good queries. An unlabeled datapoint is queried, and an optimal pruning is found by deciding if the queried node should be split – if it is, we add it to the list of pruned nodes, and if not then we recurse down the tree until we find one to add to the list. Another point is queried, and this process is repeated until the we exhaust the number of queries in our budget, each time updating the pruning list. It goes without saying that accuracy improves with an increase in the number of queries, but this framework allows us to account for the cost of those extra queries as well as design the queries we do make to optimize prediction results.
In this way we shift the transfer learning problem from a model-specific one to a model-independent one, as the HATL algorithm itself exploits similarities in data, rather than similarity in model “knowledge”, which is what traditional transfer learning approaches aim to do. This could be beneficial in reference to the problem of omic prediction posed in the last post, in which we are trying to predict gene expression in our Salmonella omic by pretraining our model with the E. coli omic. As the current model for gene expression prediction in both omics consists of only an input and output layer, it would be difficult to apply the traditional “layer swap” transfer learning. HATL also saves us from the complexities of engineered approaches.
References:
[1] David C. Kale, Marjan Ghazvininejad, Anil Ramakrishna, Jingrui He, and Yan Liu. Hierarchical active transfer learning. In Proceedings of the SIAM International Conference on Data Mining, pages 514–522, 2015.