Challenges in transfer learning in biological data
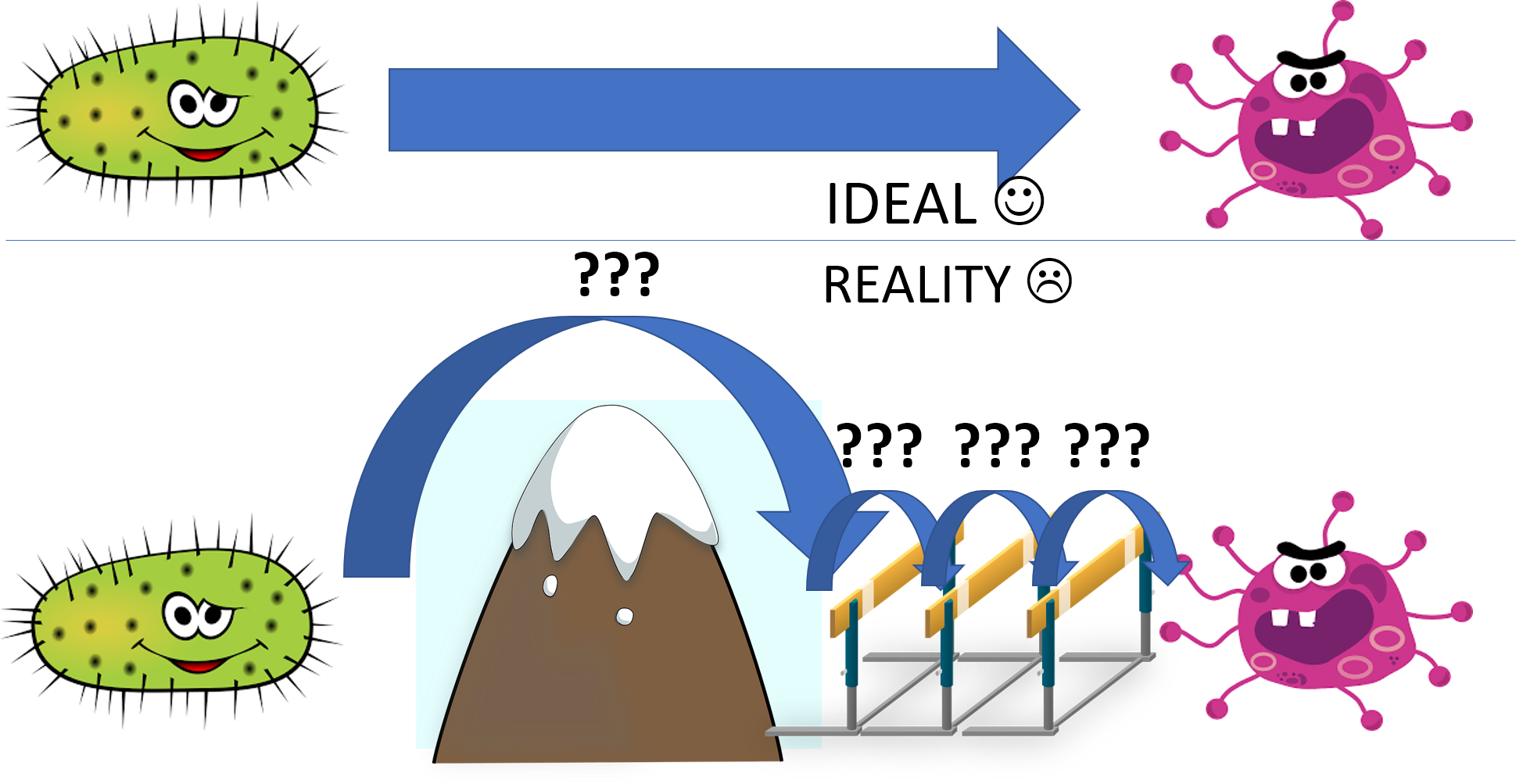
A well-defined transfer learning approach is desirable in computational biology field, as it allows us to learn better in a specific field by utilizing the knowledge in related fields. However, this approach is not trivial at all, and so far there are some notable challenges in applying transfer learning in biological data:
- Different biological features and labels among different species: For image identification, it is relatively not so difficult to apply transfer learning: The size of all images is the same, or you can resize them as you want to make the images can be fitted into the model. However, you cannot alter the features as easily in biological data. For example, E.coli contains about 4000 genes and Salmonella contains about 3000 genes, and there is no direct and reasonable way to resize 3000 features into 4000 features. Features will also be different between datasets of two differing species.
- Finding their common characteristics: Each strain or each serovar have different characteristics including the specificity of host [1]. For example, Salmonella choleraesuis infects pigs specifically but not other animals [2]. The exact mechanisms to describe the difference among strains or serovars are not clear, which increase the difficulties in finding their common characteristics. The unknown strain characteristics between two different species (e.g. E.coli and Salmonella) make transfer learning much more difficult.
There are few ways we are trying to solve these problems:
- Finding the homologous genes: We can find homologous genes between two different species, with the expectation that they share the same characteristics, including abnormal condition responses and expressions
- Focusing on the base/simpler case: To address the second issue, finding a base case is important for finding their common characteristics. If we can find the wild-type strain in both species and obtain the corresponding wild-type data, we can use this as a base for comparison. It is obviously more desirable if we have data for two species cultured in the same abnormal conditions.
References: