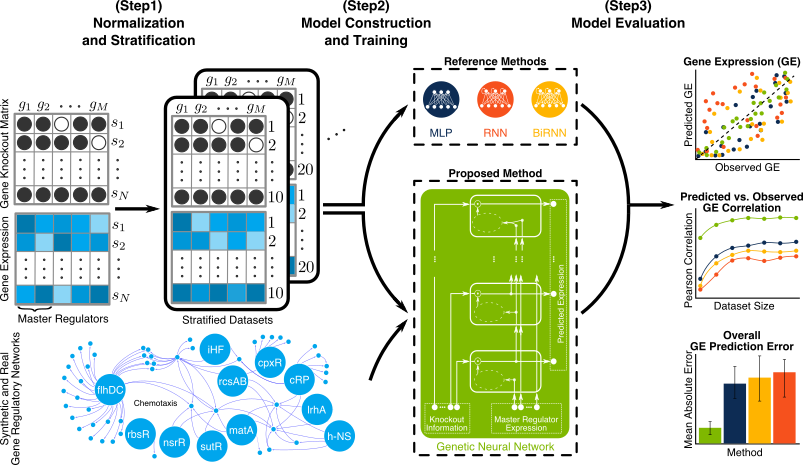
Sensible Machine Learning
There are many choices and assumptions to make when designing a machine learning (ML) based system. Taking the common choice is appealing but can undermine your system performance. Having recently designed an ML based system for prediction of gene expression (GE) [1], we made various uncommon but sensible choices and assumptions given the particular problem we solved. I’d like to highlight some of those choices here and elaborate why they are sensible. Figure 1. briefly introduces our GE prediction problem where we want our model to use expression of master regulator (MR) gene and knockout vector to predict GE values for other genes.
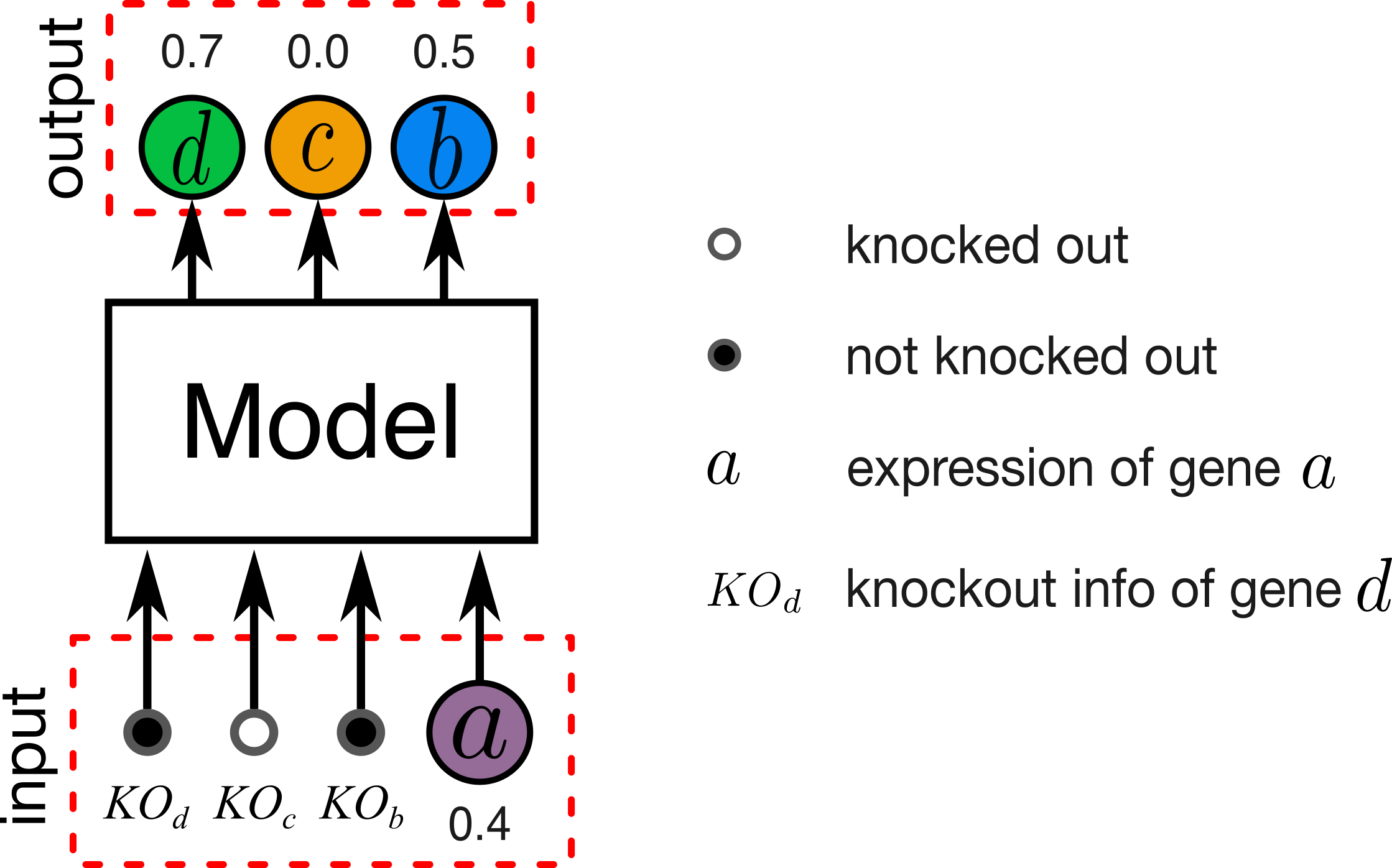
Figure 1. Illustration of gene expression prediction problem. In our experiments a given model involves between 10 to 1000 genes.
Choice of Model
We used the framework of artificial neural networks (ANNs) because of their capability to model complex relationships and create diverse architectures. This allowed us to incorporate gene regulatory network information into the model and create a new architecture we call Genetic Neural Network (GNN). Other common ANN architectures and linear models did not perform well.
Assumption of Input-Output Dependencies
When designing a predictor with multiple outputs, it is common for each output to be predicted independently of others. However it is well known in biology that genes can regulate the expression of each other hence their expression can be dependent. Our architecture incorporates such dependencies into the GNN’s architecture. Furthermore we connect each knockout signal to its related gene only.
Choice of Activation Function
Common activation functions in ANNs include Sigmoid, Tanh, ReLU and many others. We designed a new activation function based on the generalized logistic function that recapitulates the non-linear dynamics that govern gene expression.
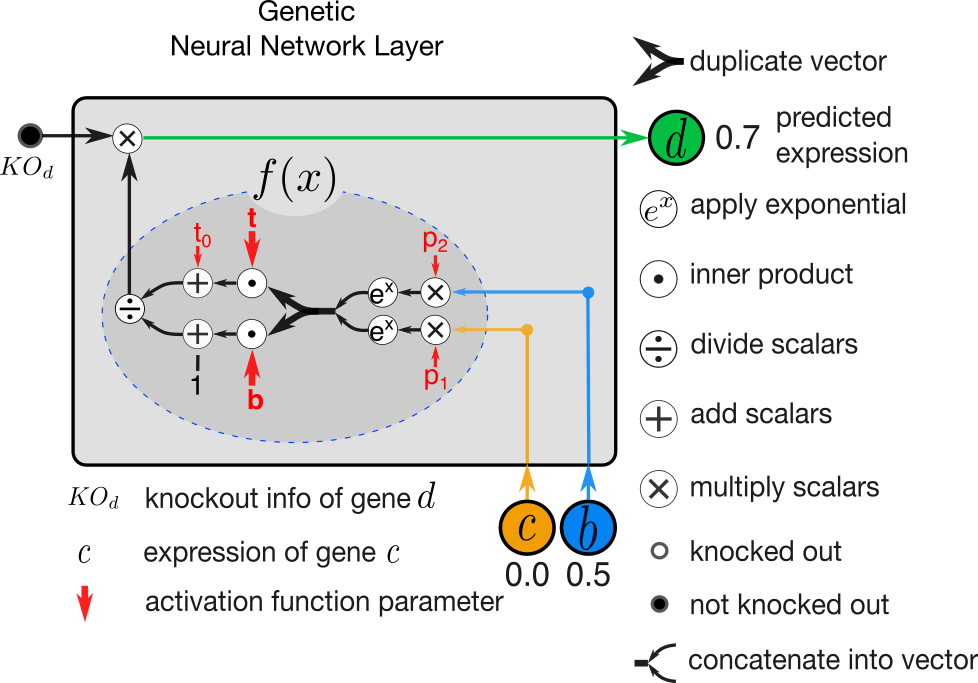
Figure 2. A GNN layer illustrated to predict expression of gene d when regulated by genes c and b.
Assumption of Parameter Dependencies
When fitting model parameters, it is common to update all parameters simultaneously in each step of optimization. We depart from this practice in two ways. First, we perform layer-wise training where each layer of GNN is trained independently. Second, in training of each layer we identify two groups of parameters where the values of the second group is dependent of the first group and achieved using a linear program. In activation function illustrated in Figure 2., the parameters t and b are dependent on p through a linear program. For detailed explanation see section “2.2 Layer-wise Training” in [1].
References
[1] A. Eetemadi and I. Tagkopoulos, “Genetic Neural Networks: An artificial neural network architecture for capturing gene expression relationships,” Bioinformatics, Nov. 2018, Advance online publication. doi: 10.1093/bioinformatics/bty945